[ad_1]
Ethics
Human tissue and clinical trials
All tissues used for isolation were obtained with informed consent and conformance to HIPAA regulations to protect the privacy of the donor’s personally identifiable information. Humans who participated in any clinical trial in this study provided written consent for study participation. The entirety of this study adheres to the Declaration of Helsinki.
Phase 0 clinical trial in Australia
This trial was conducted at the CMAX Clinical Research Center, 18A North Terrace, Adelaide, SA 5000, Australia. This phase 0 clinical trial, also the first-in-human clinical trial of INS018_055, was conducted in Australia (ACTRN12621001541897). This trial was conducted in accordance with the ethical principles of good clinical practice, according to the International Council for Harmonisation (ICH) Harmonised Guideline E6(R2) Integrated Addendum to ICH E6(R1): Guideline for Good Clinical Practice ICH E6(R2), annotated with comments by the Australian Therapeutic Goods Administration (2018). This study was approved by the Bellberry Human Research Ethics Committee.
Phase I clinical trial in China
This trial was conducted at Zhejiang Xiaoshan Hospital, 728 Yucai North Road, Xiaoshan District, Hangzhou, Zhejiang 311202, China. Detailed information including facilities and inclusion and exclusion criteria can be found at the following link: http://www.chinadrugtrials.org.cn/clinicaltrials.prosearch.dhtml (registration number CTR20221542). This study was approved by the Zhejiang Xiaoshan Hospital clinical trial ethics committee. The number of the ethics committee approval letter is EC-2022102505.
Phase I clinical trial in New Zealand
This trial was conducted at the New Zealand Clinical Research Center, level 4, 264 Antigua Street, Central City, Christchurch, 8011, New Zealand (registration number NCT05154240). This study was approved by the Northern B Health and Disability Ethics Committee. The ethics reference number for this protocol is 2021 FULL 11770.
Animal studies
All animal studies were ethically and humanely conducted following institutional animal care and use committee (IACUC), IRB or relevant animal-handling ethics organizational guidelines by our partnering CROs. Murine LPS and bleomycin studies were carried out by HD Biosciences, which were approved under the following AUF protocol numbers: AUF 146 and AUF 117. These AUF protocols were approved by the IACUC at HD Biosciences, and the studies were conducted at an AAALAC-accredited facility. The bleomycin fibrosis study in rats that were administered INS018_055 as an inhalable agent was carried out by JOINN Laboratories. This study’s IACUC-approved procedural number is S-ACU22-1144. JOINN Laboratories is fully accredited by the AAALAC, and the study adheres to regulations and rules laid out by the IACUC. The UUO kidney study was performed by SMC Laboratories. The animal care and use committee-approved protocol number is U32. All animals used in this study were housed and cared for in accordance with the Japanese Pharmacological Society Guidelines for Animal Use. The skin fibrosis model was carried out by TheraIndx LifeSciences. This study was performed using protocols approved by the Institutional Animals Ethics Committee of the test facility, which was designed under CPCSEA guidelines for animal care. The registration number for this protocol is 1852/PO/Rc/S/16/CPCSEA. For all aforementioned studies, animals were housed in groups (<5 per cage) in temperature-controlled facilities (20–26 °C) with 12-h light–12-h dark–light cycles. All animals had ad libitum access to drinking food and water.
ChatPaperGPT tool
To encourage a thorough, unbiased interrogation of our study, we developed an interactive generative AI tool, ChatPaperGPT, to navigate all contents of our study. We encourage all readers to critically examine our study, its accompanying state-of-the-art drug-discovery methodology and all associated data using our ChatPaperGPT tool (https://papers.insilicogpt.com/).
Antibodies
Antibodies used were anti-human-fibronectin (Invitrogen, MA5-11981), anti-human E-cadherin (BD Biosciences, 610182), anti-human N-cadherin (D4R1H) (Cell Signaling, 13116S), anti-human phospho-SMAD2 (Ser465/467)/SMAD3 (Ser423/425) (D27F4) (Cell Signaling, 8828S), anti-human phospho-SMAD2 (Ser465/467) (Cell Signaling, 3108S), anti-human SMAD2/SMAD3 (D7G7) (Cell Signaling, 12470S), anti-human phospho-FAK (Tyr397) (D20B1) (Cell Signaling, 8556), anti-human FAK (Cell Signaling, 3285), anti-human TNIK (Cell Signaling, 32712), anti-human NF-κB p65 (D14E12) (Cell Signaling, 8242S), anti-human phospho-NF-κB p65 (Ser536) (93H1) (Cell Signaling, 3033S), anti-human β-catenin (Cell Signaling, 9562), anti-human α-tubulin (Abcam, ab18251), anti-human HDAC2 (Abcam, ab12169), anti-human histone 3 (Abcam, ab176842), anti-α-SMA antibodies (Cell Signaling, 19245; Abcam, ab5694; Abcam, ab32575), anti-GAPDH (Absin, 016D), anti-collagen I antibody (Abcam, ab34710; LSL), goat anti-mouse IgG (Abcam, ab205719), goat anti-rabbit IgG (Abcam, ab205718) and goat polyclonal antibodies conjugated to HRP (DAKO, K4003; Vector Lab, PI-1000).
Enzyme-linked immunosorbent assay kits
ELISA kits used were as follows: IL-1β (Sinobest Bio, YX-E01291M), IL-4 (Sinobest Bio, YX-E00064M), IL-6 (Sinobest Bio, YX-E00066M), TNF-α (Sinobest Bio, YX-E00104M), fibronectin (Takara, MK115), procollagen type I C peptide (Takara, MK101), rat hydroxyproline (KinesisDx, K11-0512), collagen (Abcam, ab222942).
Other reagents
Other reagents used were as follows: 20× TBS Tween-20 (Thermo, 228360), 4–20% Criterion TGX Precast Gels, 26 well, (Bio-Rad, 5675671095), 4% Paraformaldehyde Fix Solution (Beyotime, P0099), bovine pituitary extract (ScienCell, 0713-1100mg), BSA (Sigma, B2064), bleomycin sulfate (MCE, HY-17565), CellTiter-Glo Buffer (Promega, G756B), CellTiter-Glo Substrate (Promega, G755B), cOmplete, EDTA-free Protease Inhibitor Cocktail (Roche, 4693134693132001), Difco skim milk (BD, 23232100), dimethylsulfoxide (DMSO) (Sigma, D2650-1100mL), DMEM high-glucose culture medium (Gibco, 8128120212), DMEM (Gibco, 119611960051), Dulbecco’s phosphate-buffered saline (DPBS) (Corning, 2103121031-CVC), eosin Y (Baso, BA4022), FBS (ExCell Bio, 12040), hematoxylin solution (Baso, BA4041), human recombinant epidermal growth factor (EGF) (Gibco, PHG0314), human TNF-α (Sigma, T0157), iBlot 2 NC Regular Stacks (Invitrogen, IB223001), keratinocyte serum-free medium (K-SFM) (Gibco, 10724011), Lipofectamine 3000 Transfection Kit (Thermo Fisher, L3000008), LPS from Escherichia coli O55:B5 (Sigma, L2880), MEM non-essential amino acids (100×) (Gibco, 11140050), methylcellulose (Sigma, M7140), MOPS SDS Running Buffer (1×) (Invitrogen, NP0001), nintedanib esylate (DC, DC8608), Novex ECL Chemiluminescent Substrate Reagent Kit (Invitrogen, WP220005), NuPAGE 4–12% Bis-Tris Gel, 15 well (Invitrogen, NP0330336BOX), NuPAGE LDS sample buffer (4×) (Invitrogen, NP0007), Opti-MEM (Gibco, 319831985062), PageRuler Plus Prestained Protein Ladder (Thermo Fisher, 226619), penicillin–streptomycin solution (10,000 U ml−1 penicillin, 10,000 μg ml−1 streptomycin) (HyClone, SV330010), Phosphatase Inhibitor Cocktail 2 (Sigma, P57265ML), Phosphatase Inhibitor Cocktail 3 (Sigma, P00445ML), PhosSTOP phosphatase inhibitor cocktail (Roche, 4906844906845001), Pierce western blot transfer buffer (10×) (Thermo Fisher, 335040), Pierce BCA Protein Assay Kit (Pierce, 223227), PMSF protease inhibitor (Dalian Meilun, MA0001-May06E), PMSF protease inhibitor (Beyotime, 105212110521210126), Recombinant Human TGF-β 1 Protein (MCE, AV6120051, R&D, 240-B-010 and 240-b002), Restore Western Blot Stripping Buffer (Pierce, 221059), radioimmunoprecipitation assay (RIPA) buffer (Sigma, R0278), RIPA lysis buffer (Beyotime, 926192092619200827), RPMI 1640 medium (Gibco, 22400089), saline (Hualu, NMPN 37022750), Subcellular Protein Fractionation Kit for Cultured Cells (Thermo Fisher, 778840), SuperSignal West Femto Maximum Sensitivity Substrate (Thermo Fisher, 334096), Thermo Scientific PageRuler Prestained Protein Ladder (Thermo, 226619), trypsin-EDTA (0.25%), phenol red (Gibco, 25200072).
Compounds
INS018_055 was provided by WuXi AppTec. SB525334, was purchased from TCI. Nintedanib esylate was purchased from DC (DC8608). Dexamethasone was purchased from Sigma (4902). Pirfenidone was purchased from DC (DC8792).
Synthesis of INS018_055
Synthesis, stability and characterization of INS018_055 are laid out in detail in Supplementary Information 4.
In vitro experiments
Cell lines
The human fetal lung fibroblast cell line MRC-5 was purchased from ATCC (CCL-171) (the cell viability assay was performed at WuXi AppTec) and Shanghai Cell Bank, Chinese Academy of Sciences (the α-SMA assay was performed at Boji). Human primary bronchial epithelial cells were derived from three donors with IPF (IPF05, IPF06 and IPF08) and three healthy donors (Br285, Br311 and 410955). Human primary lung fibroblasts were derived from three donors with IPF (IPF05, IPF06 and IPF08) and three healthy donors (FB218, 03HF67101 and FB2382). The human lung adenocarcinoma cell line A549 was purchased from ATCC (CCL-185). Human embryonic kidney 293T/17 cells were purchased from ATCC (CRL-11268). The human kidney cell line HK-2 was purchased from ATCC (CRL-2190). NHDFs were purchased from Bioalternatives (PF2).
Mycoplasma testing
All cell lines used were checked and cleared for mycoplasma at the following contract research organizations: WuXi AppTec (China), Charles Rivers (Netherlands), Guangzhou Boji Medical Biotechnological (China), Bioalternatives (France).
Cell culture conditions
All cell lines were cultured at 37 °C in a humidified atmosphere of 95% air and 5% CO2. MRC-5 cells were cultured in MEM medium supplemented with 1% GlutaMAX, 10% FBS, 1% non-essential amino acids and 1% penicillin–streptomycin (100 U ml−1). Human primary lung fibroblasts and human primary bronchial epithelial cells were cultured according to Charles River’s internal protocol. A549 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics (penicillin and streptomycin, 100 U ml−1 each). The 293T/17 cells were cultured in DMEM medium supplemented with 10% FBS and 1% penicillin–streptomycin solution (100 U ml−1). HK-2 cells were cultured in K-SFM containing 50% growth factor. Human primary lung fibroblasts, NHDF cells, were cultured in DMEM supplemented with 2 mM glutamine, 50 U ml−1 penicillin, 50 μg ml−1 streptomycin and 10% fetal calf serum.
Surface plasmon resonance assay for binding to His-tagged TNIK (9–315)
Binding kinetics for INS018_055, NCB-0846 and KY-05009 were measured with the Biacore 8K system. His-tagged TNIK (9–315) (WuXi AppTec) was immobilized on an NTA sensor chip for 43 s at 30 µg ml−1 and 5 µg ml−1 (His capture and amine coupling). The three compounds INS018_055, NCB-0846 and KY-05009 were prepared by twofold dilutions from 1,000 nM to 2 nM. The association time and the dissociation time were 90 s and 180 s, respectively (flow rate, 30 μl per min; sample compartment temperature, 15 °C; analysis temperature, 15 °C). The running buffer contained 20 mM HEPES, pH 7.5, 150 mM NaCl, 1 mM MgCl2, 0.05% Tween-20, 2% glycerol, 1 mM TCEP and 2% DMSO.
Enzyme selectivity profiling
INS018_055 was tested against selected kinases using Eurofins standard KinaseProfiler assays. A total of 430 kinases were tested. Protein kinases (with the exception of ATM(h) and DNA-PK(h)) were assayed in a radiometric format, whereas lipid kinases, ATM(h), ATR/ATRIP(h) and DNA-PK(h) were assayed using a homogeneous time-resolved fluorescence format. INS018_055 was prepared in a working stock 50× the final assay concentration in 100% DMSO. The required volume of the 50× stock of INS018_055 was added to the assay well, and then a reaction mix containing enzyme and substrate was added to the well. ATP at the selected concentration initiated the reaction. Data were processed by custom-built in-house analysis software at Eurofins. Results are presented as percentages of kinase activities remaining in comparison to the DMSO control. The following formula was used to calculate these values:
(Mean of sample counts − mean of blank counts)/(mean of control counts).
IC50 values were calculated using XLfit version 5.3 (ID Business Solutions). Sigmoidal dose–response (variable slope) curves were fit based on the mean result for each test concentration. Non-linear regression analysis was applied. When the top or bottom of the curve falls >10% outside of 100 or 0, either or both of these limits may be constrained at 100 and 0 when the QC criterion on R2 is met.
Measurement of FMT and EMT on human primary cells by high-content analysis
For the measurement of FMT by α-SMA, on the first day, human lung-derived primary lung fibroblast cells were seeded. After 2 d, the medium of the cells was refreshed. On day 5, cells from donors with IPF or healthy donors were prepared in two sets and exposed to INS018_055. After 1 h, all cells were treated with 1.25 ng ml−1 TGF-β1. On the eighth day (72 h after triggering), cells were fixed with 4% formaldehyde, stained for α-SMA and with DAPI and then imaged and quantified via high-content analysis (HCA) (IN Cell Analyzer 2200, GE Healthcare).
For the measurement of EMT by FN1, on the first day, human primary bronchial epithelial cells were seeded. After 2 d, the medium of the cells was refreshed. On day 6, cells from donors with IPF or healthy donors were prepared in two sets and exposed to INS018_055. After 1 h, all cells were treated with 5 ng ml−1 TGF-β1. On the ninth day (72 h after triggering), cells were fixed with 4% formaldehyde, stained for FN1 and with DAPI and then imaged and quantified via HCA (IN Cell Analyzer 2200, GE Healthcare).
Analysis of α-SMA and FN1
Segmentation and quantification of α-SMA and FN1 immunoreactivity was carried out with an HCA algorithm, with density × area (D × A) output. Data normalization of raw α-SMA or FN1 (D × A) to percent inhibition values was performed on a plate-to-plate basis.
Percent inhibition = (100 − (μp − Xi)/(μp − Xn)) × 100
μp is the average α-SMA or FN1 value of the positive control (TGF-β1 + 1 μM SB525334). μn is the average α-SMA or FN1 value of the vehicle control (TGF-β1 + 0.1% DMSO). Xi is the compound α-SMA or FN1 value. IC50 values for all compounds (if calculable, based on the point of inflection) were calculated with GraphPad Prism using non-linear fit of log (inhibitor) versus response (four parameter).
Analysis of percent remaining cells
DAPI fluorescence was applied for HCA-based quantification of the number of imaged cells on a plate-to-plate basis.
Percent remaining cells = (Xi/μn) × 100%
μn is the average number of nuclei of the vehicle control (TGF-β1 + 0.1% DMSO). Xi is the compound number of nuclei.
INS018_055 treatment of A549 cells in the TGF-β- and TNF-α-induced EMT assay
A549 cells were cultured in RPMI 1640 medium supplemented with 10% FBS and antibiotics (penicillin and streptomycin, 100 U ml−1 each) at 37 °C in a humidified atmosphere of 95% air and 5% CO2. A549 cells were seeded into six-well plates at 4 × 105 cells per well. After overnight culture, the medium was replaced with serum-free medium for 24 h of starvation before cellular induction with TGF-β alone at a final concentration of 5 ng ml−1 or with TGF-β and TNF-α at a final concentration of 20 ng ml−1 in combination with INS018_055 or DMSO for an additional 48 h, respectively. At the end of induction and compound treatment, whole-cell lysates were prepared using RIPA buffer (Sigma, R0278) with protease inhibitor or nuclear and cytoplasm fractions were extracted using the Subcellular Protein Fractionation Kit (Thermo Fisher Scientific).
Knockdown of TNIK by short hairpin RNA in A549 cells
TNIK short hairpin RNA sequences and vector information
TNIK shRNA79 was cloned into the vector PLKO.1-puro by a CRO (GENEWIZ). Packaging plasmids (psPAX2, PCMV-VSV-G) were purchased from Addgene. Sequences were as follows: shRNA control (F, 5′-CCGGCAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTGTTTTT-3′; R, 5′-AATTAAAAACAACAAGATGAAGAGCACCAACTCGAGTTGGTGCTCTTCATCTTGTTG-3′), shTNIK-1 (F, 5′-CCGGGCTCCTAAACCGTATCATAAACTCGAGTTTATGATACGGTTTAGGAGCTTTTT-3′; R, 5′-AATTAAAAAGCTCCTAAACCGTATCATAAACTCGAGTTTATGATACGGTTTAGGAGC-3′), shTNIK-4 (F, 5′-CCGGGGGCAAGGCAAAGTCTATAATCTCGAGATTATAGACTTTGCCTTGCCCTTTTTG-3′; R, 5′-AATTCAAAAAGGGCAAGGCAAAGTCTATAATCTCGAGATTATAGACTTTGCCTTGCCC-3′).
Lentivirus packaging
Lentivirus were produced in 293T/17 cells transfected with PLKO.1-puro-shRNA (shRNA control, shTNIK-1 and shTNIK-4) using the Lipofectamine 3000 transfection kit. Next, 293T/17 cells were seeded in DMEM complete culture medium in a 10-cm tissue culture dish. Cells were incubated at 37 °C with 5% CO2 overnight. Mixture I was composed of 5 µg PLKO.1-puro-shRNA, 7.5 µg psPAX2, 2.5 µg PCMV-VSV-G, 30 µl P3000 reagent and 1,500 µl Opti-MEM. Mixture II was composed of 45 µl Lipofectamine 3000 and 1,500 μl Opti-MEM.
After a 5-min incubation at room temperature, mixture I and mixture II were mixed and incubated for another 20 min at room temperature to allow DNA–Lipofectamine 3000 complexes to form. DNA–Lipofectamine 3000 complexes were added to the 293T/17 culture plate and incubated overnight. The next day, medium containing the DNA–Lipofectamine 3000 complexes was removed and replaced with fresh culture medium. Cells were incubated at 37 °C for another 72 h in a CO2 incubator.
At 72 h after transfection, virus-containing supernatants were collected and filtered with a 0.45-µm filter. Viral stocks were aliquoted and stored at −80 °C.
Infection
RPMI 1640 medium supplement with 10% FBS and 1% penicillin–streptomycin was used for A549 culture medium. A549 cells were seeded at 2 × 106 cells per plate in culture medium in a 10-cm dish and incubated with 5% CO2 at 37 °C overnight. The next day, the complete culture medium was replaced with fresh medium containing different virus ratios of 1/10 and 1/20. Virus at each ratio was used to treat cells in each dish. A total of 10 ml medium containing virus was added to each dish. Polybrene was added to each dish at a concentration of 6 μg ml−1, and dishes were incubated overnight. The next day, the medium was replaced with fresh culture medium, and the cells were incubated for another 48 h.
TGF-β treatment
After 2 d of incubation, the infected A549 cells were split into six-well plates at a density of 4.0 × 105 cells per well in culture medium and cultured overnight. The next day, the medium was substituted with a serum-free medium for 24 h of starvation. A549 cells were treated in serum-free medium with or without 5 ng ml−1 TGF-β in duplicate for 48 h at 37 °C after starvation. At the end of induction, cells were lysed in RIPA buffer supplemented with protease inhibitor and Phosphatase Inhibitor Cocktails 2 and 3 (Sigma). The supernatant was stored at −80 °C after centrifugation. The total amount of protein in the lysate was determined using the BCA Protein Assay Kit. In experiment I, cell morphology was captured with a ×4 objective lens after induction and finally was magnified at ×40.
INS018_055 treatment of MRC-5 cells
MRC-5 cells were cultured in a monolayer with DMEM high-glucose medium supplemented with 10% FBS at 37 °C with 5% CO2. Cells were seeded in six-well plates in three replicate wells per condition at a density of 2.5 × 105 cells per well and cultured for 24 h. The medium was replaced with serum-free medium, and cells were starved for 4 h. Afterward, the compound at eight concentrations (~100 μM–1.28 nM, fivefold gradient dilution) was added into each well. Vehicle (2% FBS–DMEM, containing 0.1% DMSO) was added into the blank control group and the model group, and pre-incubation was continued for 2 h. Next, TGF-β1 was added into each well of the administration group and the model group (final concentration in each well was 2 ng ml−1), and the same volume of vehicle was added into each well of the blank control group for 72 h of incubation (the culture medium was changed once after 48 h of incubation). After incubation, the cells were collected and digested with 0.5% trypsin, protein was extracted and total protein content was determined by the BCA method.
INS018_055 treatment of HK-2 cells
INS018-055 stock was diluted to eight concentrations in a threefold dilution with DMSO from the final highest concentration at 10 μM. A total of 3.0 × 105 HK-2 cells per well were seeded into a six-well plate containing 2.0 ml culture medium at a density of 3.0 × 105 cells per well. The cells were cultured overnight at 37 °C. The next day, the complete medium was replaced with K-SFM medium supplemented with 0.025 mg ml−1 bovine pituitary extract, 2.5 ng ml−1 EGF and 1% penicillin–streptomycin for starvation. After 24 h of incubation, HK-2 cells were treated with compounds (eight concentrations, threefold dilution) for 30 min before being stimulated with 8 ng ml−1 TGF-β for an additional 48 h. TGF-β induction was used as the maximum induction positive control. The final DMSO concentration in the assay was 0.1%. Cells were cultured for 48 h at 37 °C. The culture medium was discarded at the end of induction. Cells were washed once using ice-cold DPBS. RIPA buffer was added to lyse the cells for 20 min. Cells were collected, and the supernatant was stored at −80 °C. The total amount of protein in the lysate was determined using the BCA Protein Assay Kit.
Protein extraction and western blot
For whole-cell lysates, cells were collected and lysed on ice for 20 min with RIPA buffer with protease and phosphatase inhibitors (cOmplete and PhosSTOP, Roche). The protein concentration of the cell lysates was determined using the BCA Protein Assay Kit. After denaturation, whole-cell lysates and cytoplasm and nuclear extraction fractions were loaded onto 4–12% gradient Bis-Tris gels (15 wells) and 4–20% Tris-glycine gels (26 wells), respectively. For western blot detection of target proteins, 15 μg of whole-cell lysates, 5–10 μg of the cytoplasm fraction, 1–2 μg of nuclear matrix and 2–4 μg of the chromatin fraction were loaded into each lane. For western blot detection of histone 3, 0.4–1 μg of the chromatin fraction was loaded. The gels were run for 0.5 h at 80 V before being run at 120 V for another 1 h. When electrophoresis was completed, the gels containing target proteins were transferred onto NC membrane using the iBlot 2 Gel Transfer Device for 13 min at 20 V. All membranes were blocked with 5% BSA in TBST buffer at room temperature for 1 h and then incubated with primary antibodies specific to the indicated targets in TBST buffer containing 5% BSA at 4 °C overnight. After the primary antibody, membranes were washed three times with TBST buffer before incubation with HRP-conjugated secondary antibodies at room temperature for 1 h. Membranes were stripped with Restore Western Blot Stripping Buffer (Thermo Fisher) for GAPDH reference protein detection after exposure for the target proteins. Images of blots were acquired using Image Quant LAS 4000. Integrated intensity of bands from 16-bit blot images was used for quantitation with the software Image Studio Lite. Protein-quantification data of two independent experiments were calculated and analyzed using GraphPad.
α-SMA evaluation in normal human dermal fibroblasts
NHDF cells were seeded in a 96-well plate and cultured for 24 h. The medium was then replaced with an assay medium containing or lacking (control conditions) the test compound, and TGF-β (0.1 ng ml−1) was added or not (unstimulated control). The cells were then incubated for 72 h. All experimental conditions were performed in five replicates. For in situ immunolabeling and image analysis, at the end of the incubation, fibroblasts were rinsed, fixed and permeabilized. The cells were then labeled using a primary antibody (anti-α-SMA), followed by the corresponding fluorescent secondary antibody (GAM Alexa 488). In parallel, cell nuclei were stained using Hoechst solution 33258 (bisbenzimide). Image acquisition was performed using a high-resolution imaging system, the IN Cell Analyzer 2200 (GE Healthcare) automated microscope (objective lens, ×20). Five pictures per well were taken. Labeling was quantified by measuring the fluorescence intensity of α-SMA signals normalized to the total number of nuclei identified (integration of numerical data was performed with the Developer Toolbox 1.5, GE Healthcare software).
Fibronectin and procollagen I evaluation in NHDFs
NHDF cells were seeded in 96-well plates and cultured for 24 h. The medium was then replaced with assay medium containing or not containing (control conditions) the test compounds, and TGF-β (10 ng ml−1) was added or not (unstimulated control). The cells were then incubated for 72 h. Fibronectin and procollagen I contents were measured in the culture supernatants using specific ELISA kits (Takara, MK115 for fibronectin and MK101 for procollagen type I) according to the supplier’s instructions.
Cell viability
MRC-5 cells were seeded at 8,000 cells per well in 10% FBS medium and cultured with concentrations diluted from 100 μM for 72 h at 37 °C with 5% CO2. A549 cells were seeded at 5,000 cells per well in 10% FBS medium and cultured with concentrations diluted from 50 μM for 72 h at 37 °C with 5% CO2. HK-2 cells were seeded at 8,000 cells per well in 10% FBS medium and cultured with concentrations diluted from 50 μM for 72 h at 37 °C with 5% CO2.
CellTiter-Glo (Promega) was added to all conditions followed by 15 min of incubation at room temperature. Luminescent signal was detected with PerkinElmer EnVision. Data analysis was performed by calculating cell viability (%) with luminescent signal: cell viability (%) = (luminescent signal of sample − mean luminescent signal of medium)/(mean luminescent signal of cell control − mean luminescent signal of medium) × 100. CC50 values were determined according to the fitting curve of the emission ratio versus log (compound concentration) with GraphPad Prism.
Animal studies
Mice
For the bleomycin-induced lung fibrosis model and the LPS-induced acute lung injury model, C57BL/6 mice (male, 8 weeks old, ~23–26 g) were purchased from Jiangsu Gempharmatech. Animals were acclimatized for 1 week before the experiment. All in vivo experimental procedures were approved by the IACUC. All euthanasia was performed using CO2 inhalation, and all efforts were made to minimize animal suffering.
For the UUO study, 7-week-old female C57BL/6 mice were obtained from Japan SLC. Animals were housed and fed a normal diet (CE-2, CLEA Japan) under controlled conditions. All animals used in the study were housed and cared for following the Japanese Pharmacological Society Guidelines for Animal Use.
Animal models
Preparation for dosing formulations for animal model studies
For bleomycin formulation for lung fibrosis, bleomycin was dissolved in saline and vortexed to obtain a clear solution. For bleomycin formulation for the scarring model, bleomycin was dissolved in PBS. For LPS formulation, LPS was dissolved in saline and vortexed to obtain a clear solution. INS018_055 (p.o.) treatment solutions were formulated in 0.5% methylcellulose, and formulations were prepared daily. INS018_055 (topical) was formulated in 15% DMSO, 41% Transcutol, 10% IPM, 8% glycerol, 8% oleic acid, 8% diisopropyl adipate, 3% Brij L4, 4% cetostearyl alcohol and 2% hydroxypropyl cellulose (wt/wt%). Nintedanib treatment solutions were formulated in 0.5% methylcellulose. Homogeneous SB525334 suspensions were prepared in 5% DMSO + 95% (0.5% HPMC and 0.1% Tween-80 in reverse-osmosis (RO) water), and formulations were freshly prepared before administration. Dexamethasone was formulated in 7.70 ml saline and kept on ice before use. Rapamycin was prepared in 0.2% sodium carboxymethyl cellulose and 0.25% polysorbate 80.
LPS-induced acute lung injury model
One day before the first experimental day, animals were grouped. All mice were anesthetized with Zoletil–xylazine (50 mpk and 10 mpk, i.p.). At 0 h, mice received LPS at a dose of 50 μl as 50 μg per mouse (groups 2–5, n = 8 per group) or 50 μl saline (group 1, n = 8 per group) by intratracheal administration. In the INS018_055 groups (groups 3 and 4), animals were treated with INS018_055 for 4 h after the LPS challenge. In the dexamethasone group (group 5), animals were treated with dexamethasone both 2 h before and 4 h after the LPS challenge. Twenty-four hours after the LPS challenge, animals were euthanized, and BALF samples were collected.
Bleomycin lung mouse model
Mice received bleomycin on day 1 at a dose of 0.66 mg per kg (equivalent to 1 U per kg), in a volume of 50 μl by intratracheal administration, or received saline (sham group). On day 7, dosing with the corresponding compounds was started. The vehicle group (group 1) was treated with 0.5% methylcellulose BID. Groups 2–4 were treated with INS018_055 at dosages of 3 mg per kg, 10 mg per kg and 30 mg per kg BID, respectively. Group 5 was treated with 60 mg per kg nintedanib QD. On days 5 and 21, lung function measurement was performed. On day 28, all animals were killed, and blood and tissue samples were collected along with BALF. The method of model development and study design of combination studies of INS018_055 on bleomycin-induced lung fibrosis mouse models were similar.
Lung function measures in the bleomycin-induced lung fibrosis mouse model
Lung function was measured according to the HDB standard protocol. Expiratory time, relaxation time, peak expiratory flow and peak inspiratory flow were collected and used to calculate Penh data.
Inhalation administration bleomycin-induced lung fibrosis rat model
On day 1 of the study, bleomycin hydrochloride was administered to 8–9-week-old Sprague Dawley rats at a final concentration of 1.5 mg ml−1 in sodium chloride solution (dosed volume at 1.0 ml per kg) by intratracheal atomization (once in the morning and once in the afternoon) to build the pulmonary fibrosis model. INS018_055 inhalation solutions were prepared at 0.1 mg ml−1, 0.3 mg ml−1, 1.0 mg ml−1 and 6.0 mg ml−1 in sodium citrate buffer.
Aerosol generation and environmental conditions
A German PARI compression atomizer (PARI TurboBOY) was set at an aerosol flow rate of 9–10 l min−1. Aerosols from INS018_055 solutions were generated in single-chamber mode. The dilution flow of aerosol was set at 0 l min−1, and the pumping-out flow from the chamber was set at 6 l min−1. The fluid rate was set at 0.5 ml min−1 for test article replenishment. Before nebulization, aerosol-generation conditions were validated three times.
In this experiment, temperature was set at 20–26 °C, humidity between 30% and 80%, O2 level ≥ 19% and CO2 level ≤ 1%. And these parameters were recorded in real time during the experiment.
Aerosols of INS018_055 solutions were sampled and analyzed for concentrations and sizes of aerosol particles. For concentration assessment, a glass fiber filter membrane (φ = 37 mm) was used to measure aerosol concentrations during drug administration from approximately day 8 to day 28. Sampling was taken at 5 min (±1 min) after drug administration from a random exposure port. Sampling flow for the solution of 0.1 mg ml−1 INS018_055 was 1 l min−1 for 10 min. Sampling flow for solutions of 0.3 mg ml−1, 1.0 mg ml−1 and 6.0 mg ml−1 INS018_055 was 1 l min−1 for 5 min. For particle size assessment, during administration on day 8, day 15, day 22 and day 28, an exposure port was selected randomly and connected with a next-generation impactor for sampling (MOC disc filter paper) for aerosol particle size measurement at a sampling flow rate of 15 l min−1 for 5 min. Aerosol particle size parameters were calculated with the system software: median mass aerodynamic diameter (MMAD), geometric standard deviation, fine particle fraction (percentage of fine particles with MMAD < 5 μm), etc. The actual delivered dose and parameters of aerosol particle size distribution are:
|
Index |
0.1 mg ml−1 |
0.3 mg ml−1 |
1.0 mg ml−1 |
6.0 mg ml−1 |
|
|
Actual delivered dose (mg/kg)a |
0.040 ± 0.008 |
0.136 ± 0.017 |
0.485 ± 0.081 |
2.575 ± 0.242 |
|
|
NGI |
MMAD (μm) |
2.747 ± 0.317 |
2.580 ± 0.077 |
2.435 ± 0.053 |
2.5175 ± 0.167 |
|
GSD |
1.314 ± 0.111 |
1.760 ± 0.036 |
1.856 ± 0.121 |
1.957 ± 0.066 |
|
|
FPF (%) |
99.935 ± 0.007 |
85.454 ± 4.675 |
80.866 ± 5.827 |
80.294 ± 4.187 |
|
Measurement of lung function in the lung fibrosis rat model
On day 29 and before euthanasia, animals were anesthetized with chloral hydrate (90 mg ml−1, 5 ml per kg, i.p.). Necks were cut longitudinally at the middle to isolate trachea, and a catheter was intubated into the trachea toward the tail direction and fixed with a suture thread. The anesthetized animals were placed into the plethysmography chamber, to which the airway was connected. The AniRes2005 animal lung function-analysis system was used to assess parameters including FVC, airway resistance and pulmonary compliance. The ventilator respiration rate was set to 65 beats per minute; the respiration ratio was set to 20:10; the negative pressure controller was set to 30 cm H2O. FVC detection was set to pressure-control mode, while pressure was set to 30 cm H2O. The startup mode was selected to automatically detect at the end of breath, and the ‘Start’ button was clicked to passively inhale and then turned to exhale after reaching the set pressure value. Next, the ‘Stop’ button was clicked to complete an FVC detection. Each animal was evaluated for FVC at least five times. The final result was the mean value after the maximum and minimum data were excluded for analysis.
Unilateral urethral obstruction model in mice
The surgery took place on two separate days. Mice were divided into two slots based on their body weight before the day of the surgery. On day 0 (day of surgery), UUO surgery was performed after administration of three types of mixed anesthetic agents (medetomidine, midazolam, butorphanol). Mice were shaved at the incision site, the abdomen was cut open, and the left ureter was exteriorized and ligated with 4-0 silk sutures at two points. Finally, the peritoneum and the skin were closed with sutures.
Group 1 was sham-operated mice, and group 2 was treated with vehicle BID. Groups 3–5 were treated with INS018_055 at a dose of 3 mg per kg, 10 mg per kg and 30 mg per kg BID, respectively. Group 6 was treated with the ALK5 inhibitor SB525334 at a dose of 100 mg per kg QD. Treatments were applied from day 0 to day 14. And animals were killed on day 14, and blood and tissue samples were collected.
Bleomycin-induced scarring model in rats
Animals were anesthetized using isoflurane. Using a 1-ml syringe containing a 27-gauge needle, 100 µl bleomycin (1 mg ml−1 in PBS) solution was injected subcutaneously into two sites on the shaved dorsal regions, QD for 4 weeks. Rats in the naive control group were injected daily with an equivalent volume of sterile PBS. Treatments, vehicle (topical), rapamycin (1.5 mg per kg, i.p.) or INS018_055 (0.05%, 0.15% or 0.45%, topical) were administered 30 min before bleomycin administration, QD for 4 weeks. After 4 weeks of treatment, animals were killed with CO2, and skin was collected.
Bronchoalveolar lavage fluid collection
Mice were first anesthetized with Zoletil–xylazine (50 mpk and 10 mpk, i.p.). Next, a 1.5–2-cm longitudinal incision was made on the ventral side of the neck to expose the trachea by blunt dissection, and curved forceps were placed underneath the trachea. A 10–12-cm piece of suture was then threaded underneath the trachea using forceps. The ends of the suture were then pulled cranially to ‘stretch’ the trachea toward the surgeon. A 20-G needle was then used to poke a hole in the trachea as close to the larynx as possible. A 22-G blunt stainless steel needle connected to PE tubing was placed in the hole and inserted approximately 5–8 mm into the trachea. The cannula was secured in the trachea by tying the suture around it with an overhand knot. A 1-ml syringe containing 0.4 ml saline was gently pushed forward and backward three times to recover BALF fluid. BALF was transferred into empty tubes and stored on ice. This process was repeated twice with 0.3 ml saline for a total volume of 1 ml.
BALF cytological analysis
Total cell numbers were counted with a standard hemocytometer by mixing 20 μl BALF with 20 μl trypan blue. Cytospins were prepared by cytocentrifugation using Cytospins at 106g for 5 min, and then smears of BALF cells were stained with Diff-Quick stain. Cell differentiation was performed by counting at least 400 cells using standard hemocytologic criteria to classify them as alveolar macrophages or monocytes, neutrophils, eosinophils or lymphocytes. The rest of the BALF fluid was centrifuged, and supernatants were collected and stored at −80 °C for cytokine, chemokine and total protein analysis.
BALF cytokine and chemokine analysis
From the collected BALF, concentrations of cytokines were measured from mice in groups 2–7 killed on day 28. Mouse BALF cytokines and chemokines were measured using the ELISA Analysis Kit (Sinobest Bio). Mouse BALF soluble collagen was measured using the Soluble Collagen kit (Biocolor). Mouse lung hydroxyproline content was measured using the Hydroxyproline Assay Kit (Njjcbio).
Kidney hydroxyproline content
Frozen kidney samples were processed by an alkaline acid hydrolysis method as follows. Kidney samples were dissolved in 2 N NaOH at 65 °C and autoclaved at 121 °C for 20 min. The lysed samples (400 μl) were acid hydrolyzed with 400 μl of 6 N HCl at 121 °C for 20 min and neutralized with 400 μl of 4 N NaOH containing 10 mg ml−1 activated carbon. Next, AC buffer (2.2 M acetic acid and 0.48 M citric acid, 400 μl) was added to the samples, followed by centrifugation to collect the supernatant. The prepared samples and standards (serial dilutions of trans-4-hydroxy-l-proline (Sigma-Aldrich) starting at 16 μg ml−1, each 400 μl) were mixed with 400 μl chloramine T solution (Nacalai Tesque) and incubated for 25 min at room temperature. The samples were then mixed with Ehrlich’s solution (400 μl) and heated at 65 °C for 20 min to develop color. After samples were cooled on ice and centrifuged to remove precipitates, the optical density of each supernatant was measured at 560 nm. Hydroxyproline concentrations were calculated from the hydroxyproline standard curve. Kidney hydroxyproline contents were expressed as μg per mg protein, and the amount of protein was determined using the BCA Protein Assay Kit (Thermo Fisher Scientific).
Hematoxylin and eosin staining and quantification (lung)
Left lung lobes were removed from each mouse and fixed in 10% NBF solution, embedded in paraffin and processed to obtain 4-μm sections for staining with H&E. H&E-stained slides were scanned using an Aperio ScanScope Model CS2 (Leica) at ×200 magnification. Images were then opened in the HALO Plus 5 workstation (version 2.3, Indica Labs) program. Using the pen annotation tool, the whole-lung section was selected as an annotation layer. The area of vessels was defined as background using the exclusion drawing tool. The total inflammation area was quantified by selecting the dark blue inflammation cells using the pen annotation tool manually. The percentage of H&E staining (inflammation area/total area of the lung) in the selected annotation was then calculated using the program.
Masson trichrome staining and quantification (lung)
Masson’s trichrome staining was conducted using a ready-to-use kit (Trichrome Stain (Masson) Kit, HT15, Sigma-Aldrich) as described by the manufacturer.
Lung sections were cut at a thickness of 4 μm, dried in an oven for 1 h and stained with M&T using our standard protocol. Briefly, sections were stained with Weigert’s iron hematoxylin working solution for 10 min, stained in Biebrich scarlet–acid fuchsin solution for 10 min, differentiated in phosphomolybdic–phosphotungstic acid solution for 5 min or until the collagen was not red, transferred to aniline blue solution, stained for 1 min and then dedifferentiated in 1% acetic acid solution. Next, sections were dehydrated and coverslipped for subsequent image analysis. For image analysis of collagen deposition, Masson’s trichrome-stained slides were scanned by using Aperio ScanScope Model CS2 (Leica) at ×200 magnification. Images were then opened with HALO. Using the pen tool, the whole left lung section was selected as an annotation layer. The area occupied by blue collagen fibers was measured using the pen tool. The percentage of fibrosis (positive areas) in the selected annotation was then calculated with the program. Fibrosis was expressed as a percentage per lung section.
Sirius red staining and quantification (kidney)
At the end of the UUO study, the left kidney was fixed in Bouin’s solution and embedded in paraffin. Kidney sections were stained using picro-Sirius red solution (Waldeck). To quantify the interstitial fibrosis area, brightfield images in the corticomedullary region were captured using a digital camera (DFC295) at 200-fold magnification, and the positive areas in five fields per section were measured using ImageJ software (National Institute of Health).
Immunohistochemistry staining and quantification of lung tissue
Sections (4 μm thick) were placed on slides, and, after overnight drying, paraffin was removed with xylene. Next, sections were placed in a graded ethanol series and immersed in distilled water. After heat-induced citrate antigen (pH 6.0) unmasking, sections were immersed in 3% hydrogen peroxide solution for 5 min. The sections were then incubated in blocking serum (Dako, X0909) for 15 min at room temperature, followed by using primary rabbit antibodies (anti-α-SMA and anti-collagen I) for 1 h. Next, secondary antibodies conjugated to HRP were added. For image analysis of fibrosis, stained sections were used and scanned with the Aperio CS2 Scanner machine. Images were then opened with HALO. Using the pen tool, the whole left lung section was selected as an annotation layer. The bronchus was excluded in the annotation layer. The area occupied by collagen fibers was measured using the ‘Area Quantification v2.1.3’ module. The percentage of positive areas in the selected annotation was then calculated using the program.
Immunohistochemistry staining and quantification of kidney tissue
Paraffin sections were deparaffinized and hydrophilized with xylene, 100–70% alcohol series and RO water, and then circles were drawn around the kidney sections. Endogenous peroxidase activity was blocked using 0.3% H2O2 for 5 min, and antigen retrieval was performed using antigen retrieval solution H (citrate buffer) at 121 °C for 10 min. After washing with PBS, kidney sections were treated with PBS with Tween-20 (PBST), followed by incubation with Block Ace (DS Pharma Biomedical) at room temperature for 10 min. The sections were incubated with the primary antibody (anti-collagen I) at 4 °C overnight. After washing with PBS, kidney sections were treated with PBST, followed by incubation with secondary antibody at room temperature for 30 min. After washing with PBS, kidney sections were fixed with 1% glutaraldehyde solution at room temperature for 1 min. After washing with RO water, kidney sections were treated with PBST and then colored using a chromogenic substrate (Simple Stain DAB, Nichirei Bioscience). After washing with RO water, kidney sections were immersed in an eightfold-diluted hematoxylin solution for 1 min and washed with RO water immediately. The stained sections were placed in running water for 15 min and sealed with Aquatex (Merck). For scoring of IHC analyses, brightfield images were captured using a digital camera (DFC295) at 200-fold magnification, and the score in one field per section was determined.
Evaluation of hydroxyproline and collagen content in skin
Hydroxyproline and collagen contents were measured in skin lysates using rat hydroxyproline (KinesisDx, K11-0512) and collagen (Abcam, ab222942) ELISA kits. Hydroxyproline and collagen concentrations were normalized to total protein content using the Bradford method.
Statistical analysis
For western blot figures, P values between groups were analyzed by Welch’s t-test among three independent experiments. P values < 0.05 were considered statistically significant. For LPS and bleomycin mouse models, statistical analysis was performed using ordinary one-way-ANOVA, and post hoc Šídák’s multiple-comparison test was performed to calculate statistical analysis between groups. The difference was considered significant when P < 0.05. For the inhalation study in the bleomycin rat model, in analyzing parameters of lung functions (FVC, airway resistance and pulmonary compliance), statistical analysis was performed using uncorrected Fisher’s least-significant difference test as post hoc analysis after ANOVA analysis; in analyzing pathology results, statistical analysis was performed using Kruskal–Wallis test and Dunn’s multiple-comparison test as post hoc analysis after ANOVA analysis. Differences were considered significant when P < 0.05. For the UUO model, statistical analyses were performed using the Bonferroni multiple-comparison test. P values < 0.05 were considered statistically significant. For the bleomycin-induced skin scarring experiment, statistical analysis was performed using one-way ANOVA, and then Dunnett’s multiple-comparison test was performed. P values < 0.05 were considered statistically significant. For Fig. 2h, enrichment analysis was performed using the gseapy.enrichr Python package. P values were computed using Fisher’s exact test. This is a binomial proportion test that assumes a binomial distribution and independence for probability of any gene belonging to any set. Adjusted P values (q values) were calculated using the Benjamini–Hochberg method for correction for multiple-hypothesis testing.
Single-cell RNA sequencing analysis
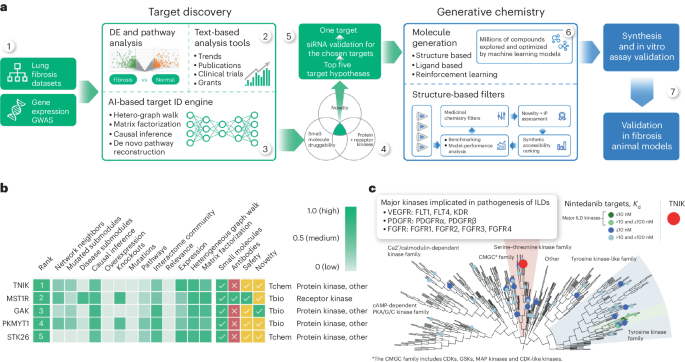
Single-cell RNA sequencing (scRNA-seq) data from 32 IPF and 28 control lungs were collected from the GEO database (GEO accession GSE136831)53. Data were preprocessed with a standard pipeline using the Scanpy package. Single-cell data was filtered by applying two conditions: including cells with a minimum of 200 genes and with genes expressed in more than three cells. Cells with mitochondrial fraction greater than 20% were excluded. Data were initially normalized with a scale factor of 10,000 and log transformed. PCA was run with the sc.pp.pca function and ‘n_comps’ = 50. Batch effects were corrected with ‘sce.pp.harmony_integrate’ function. A neighborhood graph was computed on the first 50 principal components derived after batch correction, and results were visualized using UMAP. Cell type annotation was obtained from the original article. Cluster gene signatures were identified by testing for differential expression of a subgroup against all other cells using a Wilcoxon rank-sum with the ‘tl.rank_genes_groups’ function. Differential expression between IPF and controls for each cell type was calculated using the ‘tl.rank_genes_groups function’ (method = ‘Wilcoxon’), and TNIK expression was plotted (non zero values with P values, Benjamini–Hochberg adjustment and a threshold of 0.05) using the plotly package.
Simulated knockout profile of TNIK
scTenifoldKnk54 is a method developed to perform virtual knockout experiments to predict gene functions. First, scTenifoldKnk, based on scRNA-seq data, constructs a denoised single-cell gene regulatory network (scGRN). The scGRN is copied, and then outward edges of the target gene in the adjacency matrix of the copied scGRN are zeroed out, thus creating a pseudo-knockout scGRN. Having two scGRNs (one initial net and a second pseudo-knockout net), target gene regulatory significance can be estimated with a manifold alignment procedure. Two scGRNs are mapped to the same low-dimensional space, and distance between gene projections shows the impact of gene knockout in the scGRN: larger perturbation of genes in low-dimensional space indicates the importance of the target gene in the scGRN. We used IPF myofibroblast scRNA-seq data derived from GSE136831 (ref. 53) in scTenifoldKnk and made a pseudo-knockout of TNIK. The list of perturbed genes was sorted by fold changes of distances between gene projections of two scGRNs; probabilities were assigned using χ2 distribution with one degree of freedom. The most perturbed genes are supposed to have a tight connection with the target gene. Next, we applied the MCODE algorithm47 for TNIK and the top 100 most perturbed genes and performed pathway and process enrichment analysis.
Chemistry42
The Molecular Sets platform (MOSES)87 was used to train and benchmark the generative chemistry models in Chemistry42. The structure-based drug-design workflow implemented in the Chemistry42 platform was used to generate a library of virtual structures40. Pocket and Pharmacophore Reward modules were used for scoring produced designs and navigating the generative process. The TNIK ATP-binding site was selected as a target-binding pocket. The generated structures had to match a two-point pharmacophore hypothesis that consisted of the hydrogen bond acceptor forming an H-bond with the NH of Cys108 of the hinge region and hydrophobic center occupying the space near the gatekeeper Met105. In addition to the assessment by Pocket and Pharmacophore modules, the engine penalized structures that violated the predefined ranges of the physicochemical properties (logP, molecular weight, number of hydrogen bond donors, hydrogen bond acceptor, topological polar surface area, number of atoms, number of rotatable bonds, number of aromatic rings), medicinal chemistry filters and the synthetic accessibility score threshold.
Phase 0 study (Australia)
A phase 0 micro dosing clinical trial was conducted in Australia (ACTRN12621001541897) and can be found at https://www.anzctr.org.au/TrialSearch.aspx. Complete information on clinical trial registration, study protocol, data collection and outcomes is provided in the Reporting summary as well as in Supplementary Information 8. This includes statistical considerations, study design, patient-selection criteria, procedures, outcomes and PK analysis.
Phase I study (New Zealand)
The general design of this clinical trial (NCT05154240) can be found at https://www.clinicaltrials.gov. The randomized, double-blind, placebo-controlled study of INS018_055 was conducted from 21 February 2022 (first participant administered first dose) until 30 September 2022 (last participant contacted). Complete information on clinical trial registration, study protocol, data collection and outcomes is provided in the Reporting summary as well as in Supplementary Information 9. This includes statistical considerations, study design, patient-selection criteria, procedures, outcomes and PK analysis.
Phase I study (China)
Detailed information including facilities and inclusion and exclusion criteria can be found at the following link: http://www.chinadrugtrials.org.cn/clinicaltrials.prosearch.dhtml (registration number CTR20221542).
Complete information on clinical trial registration, study protocol, data collection and outcomes are provided in the Reporting summary as well as in Supplementary Information 10. This includes statistical considerations, study design, patient-selection criteria, procedures, outcomes and PK analysis.
Full clinical study protocols for all three trials are provided in the Supplementary Information.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Maqvi News #Maqvi #Maqvinews #Maqvi_news #Maqvi#News #info@maqvi.com
[ad_2]
Source link